Work experience
Living Optics: Frontend Software Engineer; 2023.05-present
Living Optics is an optics startup spun out of research from the University of Oxford. We are trying to add value to computer vision problems using their unique hyperspectral cameras.
I joined Living Optics with the ambition of building a platform for camera control and analysis in realtime to assist users in getting the most out of their cameras.
Living optics hyperspectral camera works using a light blocking mask to let through pinholes of light. The complex optics then spreads out that light like a prism accross 96 pixels on the light receiving sensor. This data can then be reconstructed back into 96 bins, a wavelength fingerprint for that pin hole. This is repeated for around 2000 pinholes and also the spectrum for the entire field of view to be gernerated.
I have built out the frontend and backend web based camera control tool. The initial offering provides live video streaming capabilities as well as full control of the camera's settings. The backend layer I have built utilises the full power of the Living Optics core api's to handle reading, recording and reconstructing the spectral from the camera in real time at 30fps. The video feed is streamed over webrtc to one or more clients in another thread to avoid interference with the frame rate of reading and recording. Updates are made by api requests and camera state changes propagated by websocket to ensure multiple clients can be kept in sync.
The frontend is bundled and served alongside the backend using Fastapi and can be used by a remote client via the camera's wifi hotspot. The modular design of the frontend has allowed rapid redesign of the form factor of the ui without risk of introducing bugs.

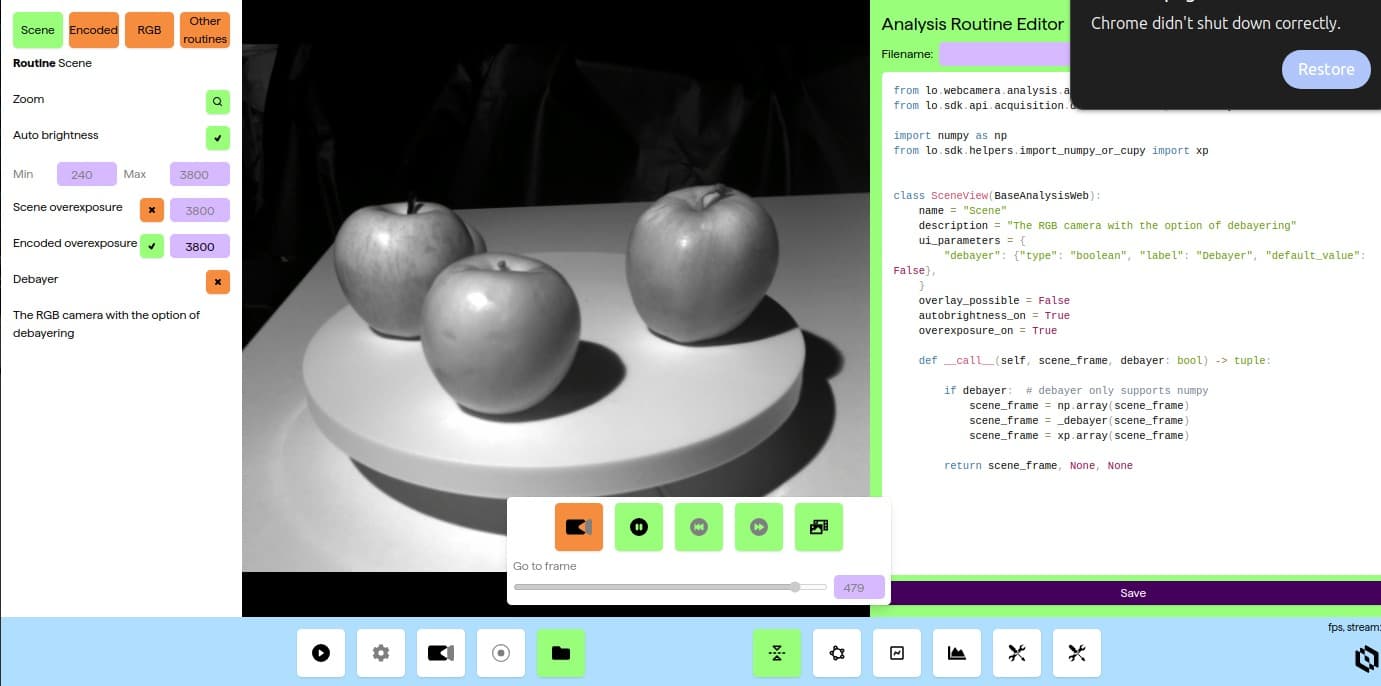
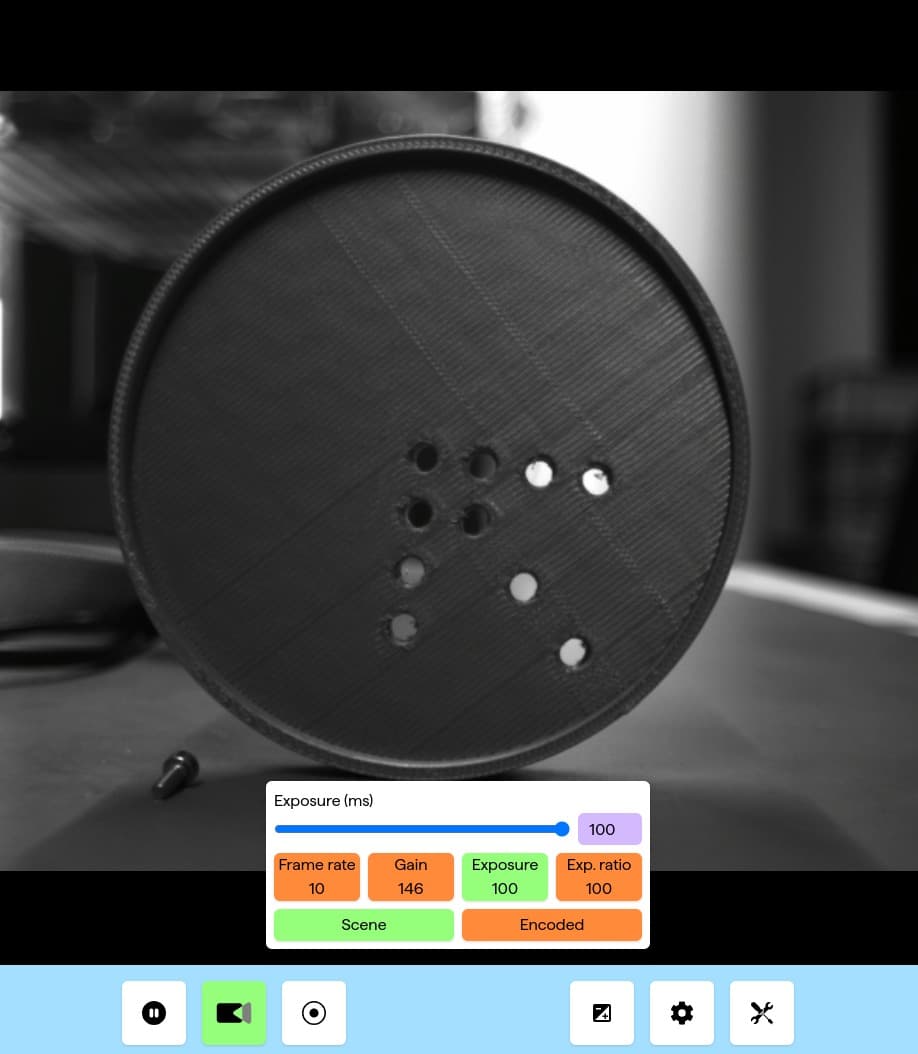
I have designed and built a realtime analysis layer on top of the base camera. This allows overlaying information on the video feed in real time from both Living Optics analysis routines and user written routines, thus making it easy for all scientist and developers to build and test new ideas. It support live editing: Code changes are applied on save making creating useful analysis routines easier.
Tech stack summary
Frontend
- function based react
- webrtc for video streaming
- redux + redux-tool-kit
- tailwindcss
- zod
- websocket communication for realtime data streaming and state updates
Backend
- Fastapi based api's
- AIORTC based video streaming
- websocket communication management, supporting multiple clients
ONI: Software Engineer; 2018.05-2023.05
ONI is a microscopy startup with the goal of making super-resolution microscopy accessible and affordable for all.
I joined ONI with the ambition to provide tools to scientists to help them better understand the data they collect.
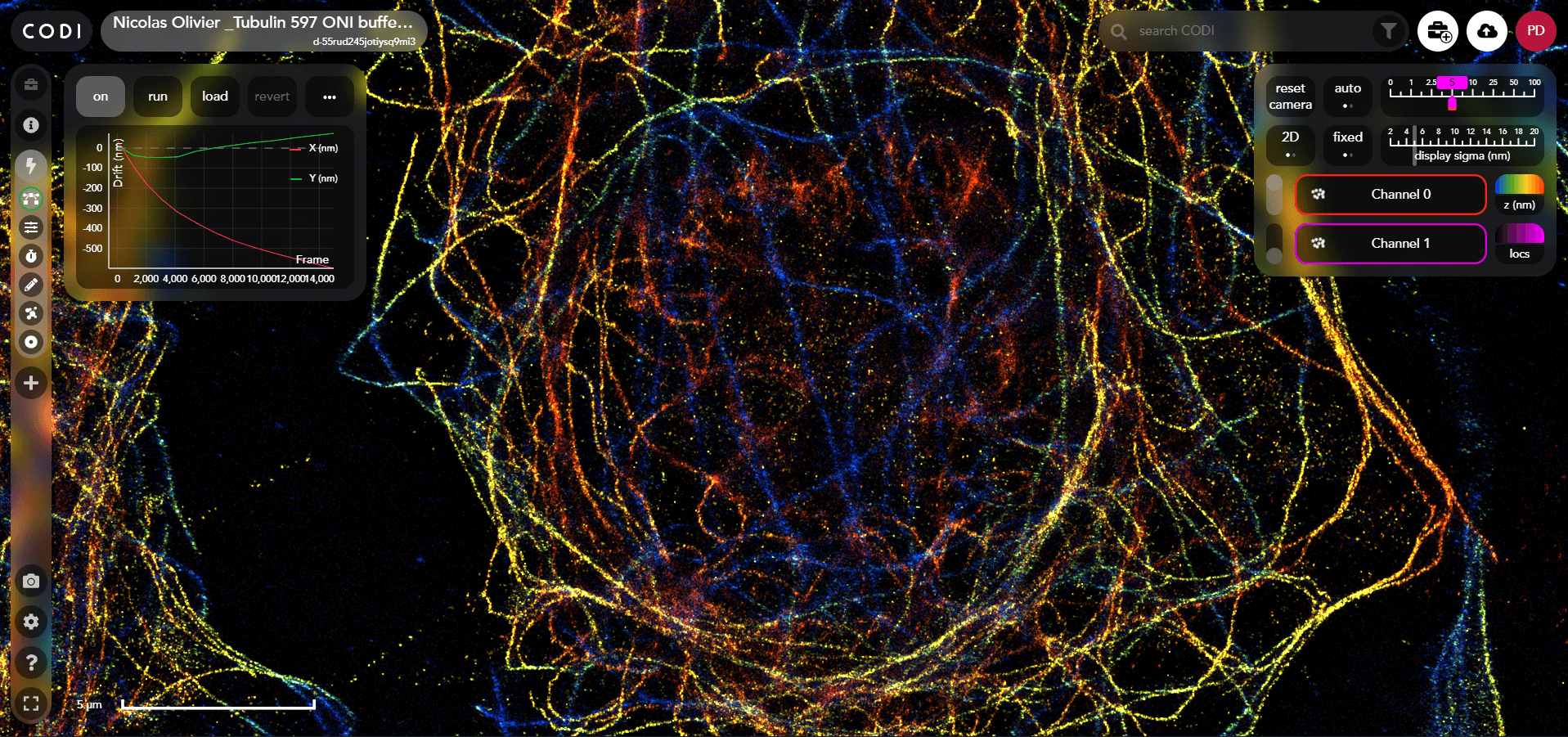
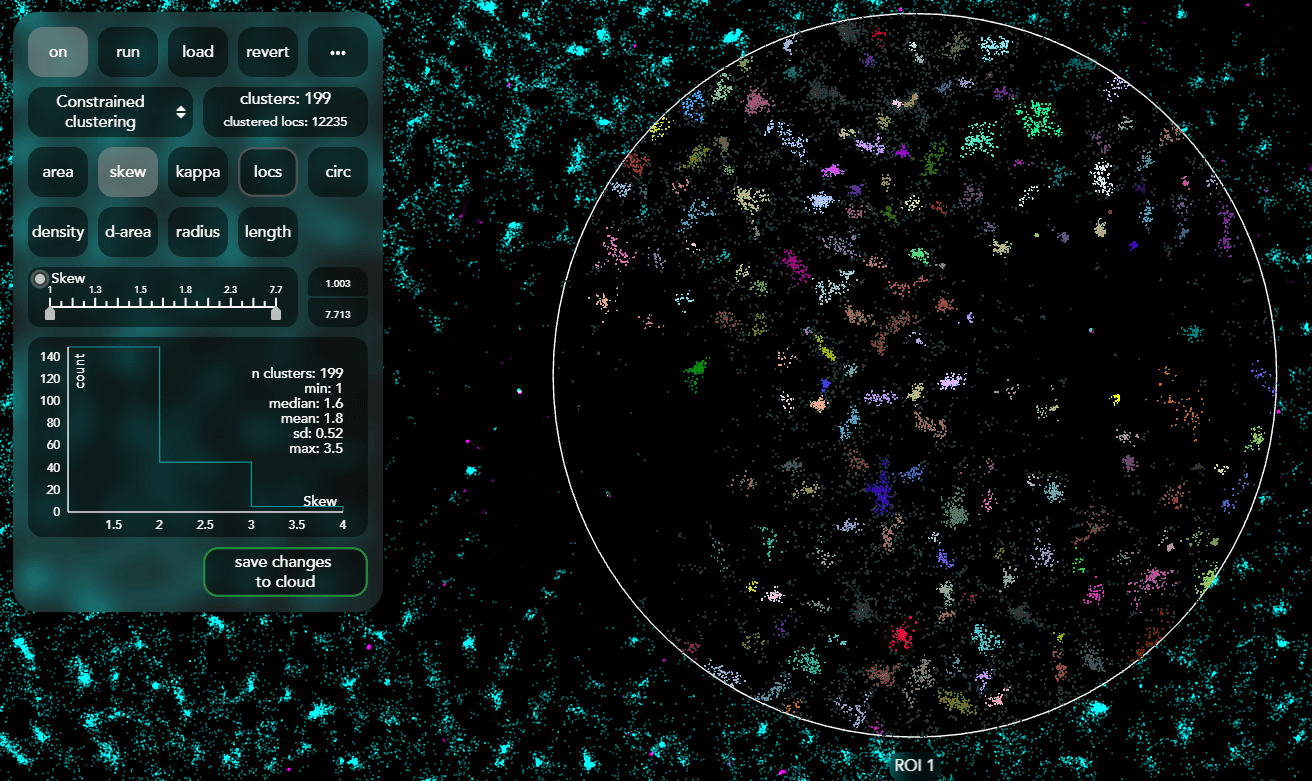
CODI is a website designed to support super-resolution imaging and analysis for data collected from ONI's microscopes. CODI allows researchers to gain insights into their data and share these insights with colleagues globally. The core team was formed in August 2018. It has been small, consisting of 4-6 people, which has given me opportunities to learn and develop skills in all aspects of web development.
I built and contributed to the vast majority of the frontend features and have helped maintain the backend and cloud infrastructure. I designed and built out front and backend infrastructure to run complex analysis pipelines. This consists of flexible steps of image analytic tools applied in any order to a dataset. This is a transition from a single hard coded pipeline to fullh composable tool.
I helped design the first version of gRPC protocols for ONI's microscope and delivered the frontend codebase. This was for a prototype web based frontend for our microscope. These consist of commands from the frontend and streams relaying changes to the microscopes state from the backend. For the prototype the microscope video feed was accessed from shared memory.
I built a big-data point cloud rendering engine in three.js which uses google maps like tiling to avoid the need to load all the data in one go whilst still providing a good representation.
I oversaw the transition from React components to functional React components and hooks whilst they were still in beta, and also the transition from React contexts to redux for global state management.
Tech stack summary
Frontend
- function based react
- redux + redux-tool-kit
- styled-components
- three.js
- gRPC + protobuf communication between frontend and microscope
Backend
- kubernetes cluster
- go server for authentication and authorization
- django server with celery workers for data analysis
Things I would change
I would adopt a common backend language. Having functionality split between two backend services causes duplication of functionality, difficulty handling permissions as well as requiring fluency in an additional language.
Asynchronous tasks run outside the kubernetes infrastructure. We currently have fixed sized celery workers sitting inside the cluster consuming resources when inactive. Since data processing tasks vary in resource demand (cpu,memory) these could be better served using an elastic compute service to ensure tasks don't bottleneck on resources. Using a serverless approach would also reduce cost through eliminating idling time.
University of Oxford, Centre for Statistics in Medicine (CMS): Trial Statistician; 2014.06-2018.05
I joined with the goal of maximising the value of every patient within a clinical trial. This focused on rare cancer studies where traditional statistical approaches are insufficient because it would take too long to recruit enough eligible patients.
I developed novel Bayesian trial designs for two bone cancer trials. Bone cancer is a rare disease which mostly affects children and young adults. The trials aimed to test the safety and efficacy of novel treatments.
- Developed a React.js based app for presenting and exploring adverse event data from clinical trials.
- Developed R shiny applications for trial design visualisations. The languages HTML, CSS, JavaScript (D3.js and JQuery) were used to complete this project.
- Authored a Bayesian sample size package in R for single-arm multi-stage phase II trials. The package is object oriented utilising strongly typed classes to streamline the functional flow and make comparing designs easy.
- Developed fully documented R packages for automated reporting and designing complex trials though simulation.
- Innovated on existing published trial designs to make them more efficient.
- Developed an R based data query system, which identifies if a problem was identified in a previous patient visit.
- Worked with complex relational databases synthesising participant trial data to produce automated reports.
- Wrote and published statistical and clinical papers in academic journals. Details here
Tech stack summary
- R and R core plotting libraries
- Shiny: an R based library for generating web apps
- d3.js
- Stata
Things I would change
Each statistician was allocated to a number of clinical trials or projects. Much work was done independently which in many cases was a waste of resources because each individual would write similar code for each trial instead of having a shared pool of utility functions to then utilise. For the most part this meant that for any new trial generating code for reports started from scratch. I did write some code with the intention of reuse when I was there and this was slowly starting to improve when I left. This also meant there was a lack of collaboration for everyday tasks.
Cardiff University, Wales Cancer Trials Unit: Trial Statistician; 2012.10-2014.06
I worked as a Trial Statistician in Cardiff supporting the reporting and statistical needs of active cancer clinical trials. I wrote statistical analysis pans, programs to reproducibly carry out the analysis and generated statistical reports. I automated frequently required reports using STATA, Visual Basic and batch files.
I also provided statistical advice to clinicians at the partner cancer hospital. This included assisting with study design and helping with analysis.
I also peer reviewed papers, for statistical quality and accuracy, for the British Medical Journal.